Machine Learning From Scratch I
Machine learning is the capability of machines to imitate human behaviour while solving problems. It involves computers using algorithms to find insightful information through an iterative process.
The machine learning process begins with inputing training data in an algorithm of choice. The training data alters the weights and biases within the algorithm, new input data is then fed into the algorithm to make predictions. The predictions are then compared with the actual values to check if the results match, if they dont. The algorithm is retrained until the desired outcome is achieved.
There are two primary areas of machine learning, the supervised and unsupervised learning.
- Supervised machine learning uses labeled data from the training data. In this piece, we build and implement supervised algorithms such as linear regression, logistic regression, decision trees, random forest, adaboost, support vector machines, k-nearest neighbors, linear discriminant analysis, and Naive Bayes.
- Unsupervised machine learning uses unlabeled data, the trained model tries to search for patterns. We explore two popular unsupervised algorithms namely, principal components analysis and K-means clustering.
Linear Regression: Least Square Method
Given independent variable and dependent variable . Using the linear model , we can calculate the slope (w) and the intercept ().
The mean of and , all the regresssion lines must go through their point of interception.
The slope
Since . Intercept
The regression formula becomes
R Squared
It tells how well a regression formula predicts the actual values, if the actual is very close to the estimates then . From our x above, we can use , with to estimate the values as follows;
Standard Error of the Estimate
The distance between the actual and the estimated (error).
Standard Deviation
The deviation of values of a variable from the mean,
Correlation
A measure of strength of association between two variables. Where as, regression predicts one variable given another variable.
0<4<0.19: v.low, 0.2<r<0.39:low, 0.4<r<0.59: moderate, 0.6<r0.79:high, 0.8<r<1: v.high
Implementing Linear Regression in Python
Use to predict continuous values
y = wx + b #w=slope, b=bias/y-intercept
Cost Function
To minimize the cost function(error), we calculate the gradient of the cost function with respect to w and b.
Gradient Descent
Initialize weight, then use alpha to move to the negative gradient until the minimum point is reached.
alpha() = learning rate(lr)
Update Rules
from sklearn import datasets
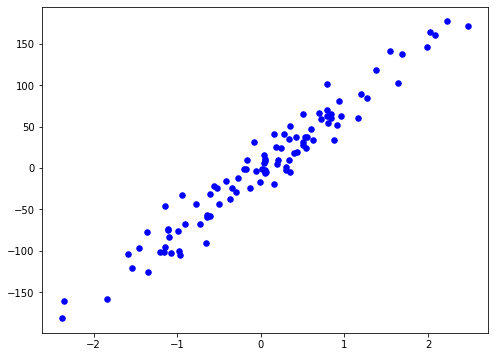
X, y = datasets.make_regression(n_samples=100, n_features=1, noise=20, random_state=5)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=5)
fig = plt.figure(figsize=(8,6))
plt.scatter(X[:, 0], y, color="b", marker="o", s=30)
plt.show()
class LinearRegression:
def __init__(self, lr=0.001, n_iters=1000):
self.lr = lr
self.n_iters = n_iters
self.weights = None
self.bias = None
def fit(self, X, y):
# init parameters
n_samples, n_features = X.shape
self.weights = np.zeros(n_features)
self.bias = 0
# Gradient Descent
for _ in range(self.n_iters):
# y = mx + c
y_predicted = np.dot(X, self.weights) + self.bias
# 2 in 2x can be ommited because it is a scaling factor
dw = (1/n_samples) + np.dot(X.T, (y_predicted - y))
db = (1/n_samples) + np.sum(y_predicted-y)
# update parameters
self.weights -= self.lr * dw
self.bias -= self.lr * db
def predict(self, X):
y_predicted = np.dot(X, self.weights) + self.bias
return y_predicted
def mse(y_true, y_predicted):
return np.mean((y_true - y_predicted)**2)
regressor = LinearRegression(lr=0.01) #0.0001
regressor.fit(X_train, y_train)
predicted = regressor.predict(X_test)
mse_value = mse(y_test, predicted)
print("Linear Regression Mean Squared Error", mse_value)
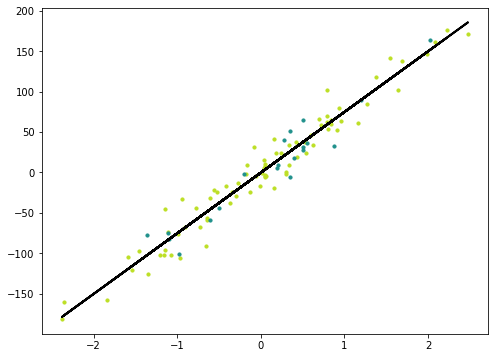
y_pred_line = regressor.predict(X)
cmap = plt.get_cmap('viridis')
fig = plt.figure(figsize=(8,6))
m1 = plt.scatter(X_train, y_train, color=cmap(0.9), s=10)
m2 = plt.scatter(X_test, y_test, color=cmap(0.5), s=10)
plt.plot(X, y_pred_line, color='black', linewidth=2, label='Prediction')
plt.show()Linear Regression Mean Squared Error 305.7734835201333Logistic Regression
Logistic Regression is used when the dependent variable(target) is categorical. Its a statistical analysis method to predict a binary outcome, such as yes or no, based on prior observations of a data set.
Approximation
Linear Function (countinuous values output)
f(w, b) = wx + b
Sigmoid Function (probabilistic output between 0-1)
Cost function
Cross entropy loss is reduced using gradient descent.
The main difference between linear and logistic regression is the sigmoid function applied to the output of a linear function
from sklearn import datasets
data = datasets.load_breast_cancer()
X, y = data.data, data.target
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=5)
class LogisticRegression:
def __init__(self, lr=0.001, n_iters=1000):
self.lr = lr
self.n_iters = n_iters
self.weights = None
self.bias = None
def fit(self, X, y):
# init parameters
n_samples, n_features = X.shape
self.weights = np.zeros(n_features)
self.bias = 0
for _ in range(self.n_iters):
linear_model = np.dot(X, self.weights) + self.bias
y_predicted = self._sigmoid(linear_model)
# update weights (2)
dw = (1/n_samples) * np.dot(X.T, (y_predicted-y))
db = (1/n_samples) * np.sum(y_predicted -y)
self.weights -= self.lr * dw
self.bias -= self.lr * db
def predict(self, X):
# using updated weights and bias
linear_model = np.dot(X, self.weights)+self.bias
y_predicted = self._sigmoid(linear_model)
y_predicted_cls = [1 if i > 0.5 else 0 for i in y_predicted]
return y_predicted_cls
# helper method
def _sigmoid(self, x):
return 1 / (1 + np.exp(-x))
def accuracy(y_true, y_pred):
acc = np.sum( (y_true == y_pred)/ len(y_true))
return acc
classifier = LogisticRegression(lr=0.0001) #0.0001
classifier.fit(X_train, y_train)
predicted = classifier.predict(X_test)
acc_value = accuracy(y_test, predicted)
print("Logistic Regression Accuracy ", acc_value)Logistic Regression Accuracy 0.9298245614035088Refactoring Regression Code
The linear and logistic regression code is similar apart from the sigmoid function transformation. We can create a base class that contains all the shared methods and properties.
class BaseRegression:
def __init__(self, lr=0.001, n_iters=1000):
self.lr = lr
self.n_iters = n_iters
self.weights = None
self.bias = None
def fit(self, X, y):
n_samples, n_features = X.shape
self.weights = np.zeros(n_features)
self.bias = 0
# Gradient Descent
for _ in range(self.n_iters):
y_predicted = self._approximation(X, self.weights, self.bias)
dw = (1/n_samples) + np.dot(X.T, (y_predicted - y))
db = (1/n_samples) + np.sum(y_predicted-y)
self.weights -= self.lr * dw
self.bias -= self.lr * db
# helper method to calculate the linear/sigmoid function value
def _approximation(self, X, w, b):
raise NotImplementedError()
# when predict method is called in base class output error
def predict(self, X):
return self._predict(X, self.weights, self.bias)
def _predict(self, X, w, b):
raise NotImplementedError()
class LinearRegressio(BaseRegression):
def _approximation(self, X, w, b):
return np.dot(X, w) + b
def _predict(self, X, w, b):
return np.dot(X, w) + b
class LogisticRegressio(BaseRegression):
# _approximation returns probabilities like predict_proba
def _approximation(self, X, w, b):
linear_model = np.dot(X, w)+ b
return self._sigmoid(linear_model)
def _predict(self, X, w, b):
linear_model = np.dot(X, w) + b
y_predicted = self._sigmoid(linear_model)
#print(y_predicted[:90])
y_predicted_cls = [1 if i > 0.5 else 0 for i in y_predicted]
return y_predicted_cls
def _sigmoid(self, x):
return 1 / (1 + np.exp(-x))
clf = LogisticRegressio(lr=0.001) #0.0001
clf.fit(X_train, y_train)
predicted = clf.predict(X_test)
acc_value = accuracy(y_test, predicted)
print("Logistic Regresssion accuracy", acc_value)Logistic Regresssion accuracy 0.9210526315789473Support Vector Machines (SVM)
SVM algorithm is mostly used for classifiction problems, we plot each data item as a point in n-dimensional space with the value of each feature being the value of a particular coordinate. In classification we find the hyper-plane that best differentiates the classes.
Uses a linear model to find a decision boundary(hyperplane) that best separates data classes. The margins between the hyperplane and the classes should be maximized (2 / ||w||).
Linear Model
Cost Function
A cost function is a formula used to predict the cost that will be experienced at a certain activity level.
Hinge Loss
Regularization
Gradients
Update Rule
For each training sample :
class SVM:
def __init__(self, lr=0.001, lambda_param=0.01, n_iters=1000):
self.lr = lr
self.lambda_param = lambda_param
self.n_iters = n_iters
self.weight = None
self.bias = None
def fit(self, X, y):
y_ = np.where(y <=0, -1, 1) #convert [0,1] to [-1, 1]
n_samples, n_features = X.shape
self.weight = np.zeros(n_features)
self.bias = 0
for _ in range(self.n_iters):
for idx, x_i in enumerate(X):
condition = y_[idx] * (np.dot(x_i, self.weight) - self.bias) >= 1
if condition:
self.weight -= self.lr * (2 * self.lambda_param * self.weight)
else:
self.weight -= self.lr * (2 * self.lambda_param*self.weight - np.dot(x_i, y_[idx]))
self.bias -= self.lr * y_[idx]
def predict(self, X):
linear_output = np.dot(X, self.weight) - self.bias
return np.sign(linear_output) # if positive: +1, if neg: -1
X, y = datasets.make_blobs(n_samples=50, n_features=2, centers=2, cluster_std=1.05, random_state=5)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=5)
y = np.where(y == 0, -1, 1)
clf = SVM()
clf.fit(X, y)
predictions = clf.predict(X)
acc = accuracy(y, predictions)
print("Svm Accuracy", acc)
print(clf.weight, clf.bias)
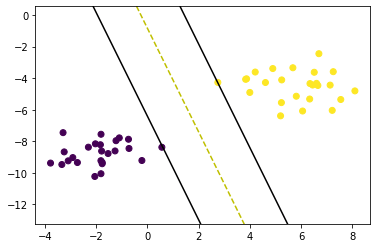
def visualize_svm():
def get_hyperplane_value(x, w, b, offset):
return (-w[0] * x + b + offset) / w[1]
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
plt.scatter(X[:,0], X[:, 1], marker='o', c=y)
x0_1 = np.amin(X[:, 0])
x0_2 = np.amax(X[:, 0])
x1_1 = get_hyperplane_value(x0_1, clf.weight, clf.bias, 0)
x1_2 = get_hyperplane_value(x0_2, clf.weight, clf.bias, 0)
x1_1_m = get_hyperplane_value(x0_1, clf.weight, clf.bias, -1)
x1_2_m = get_hyperplane_value(x0_2, clf.weight, clf.bias, -1)
x1_1_p = get_hyperplane_value(x0_1, clf.weight, clf.bias, 1)
x1_2_p = get_hyperplane_value(x0_2, clf.weight, clf.bias, 1)
ax.plot([x0_1, x0_2], [x1_1, x1_2], 'y--')
ax.plot([x0_1, x0_2], [x1_1_m, x1_2_m], 'k')
ax.plot([x0_1, x0_2], [x1_1_p, x1_2_p], 'k')
x1_min = np.amin(X[:, 1])
x1_max = np.amax(X[:, 1])
ax.set_ylim([x1_min-3, x1_max+3]) # y-limits
visualize_svm()Svm Accuracy 1.0
[0.58977016 0.17946483] -0.1520000000000001KNN
The KNN algorithm is used for classification and regression problems. It assumes that similar things exist in close proximity to each other. We calculate distances between values using Euclidean distance.
from collections import Counter
def euclidean_distance(x1, x2):
# (sum(q_i-p_i)^2)^0.5
return np.sqrt(np.sum( (x1-x2)**2))
class KNN:
def __init__(self, k=3):
self.k = k
def fit(self, X, y):
self.X_train = X
self.y_train = y
def predict(self, X):
"""
Take each test item and pass it to _predict helper
"""
predicted_labels = [self._predict(x) for x in X]
return np.array(predicted_labels)
def _predict(self, x):
"""
Calculate the pythagorean distance between each test value and
train items.
Select the k train rows with smallest distance.
Find the most common class for the train rows with smallest distance
"""
#compute distances
distances = [euclidean_distance(x, x_train) for x_train in self.X_train]
#get k nearest samples (shortest distance)
k_indices = np.argsort(distances)[:self.k]
k_nearest_labels = [self.y_train[i] for i in k_indices]
#majority vote
most_common = Counter(k_nearest_labels).most_common(1)
return most_common[0][0]
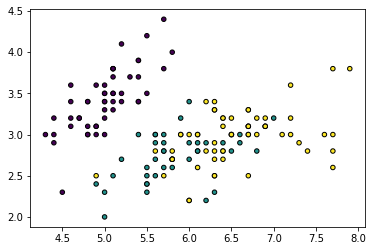
iris = datasets.load_iris()
X, y = iris.data, iris.target
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=1234)
plt.figure()
plt.scatter(X[:, 0], X[:, 1], c=y, cmap=cmap, edgecolor='k', s=20)
clf = KNN(k=2)
clf.fit(X_train, y_train)
predictions = clf.predict(X_test)
# sum of correct predictions / total test items
acc = np.sum( predictions == y_test)/len(y_test)
print("KNN accuracy", acc) # k:5 = 0.97, k:3 = 1.0, k:2=1.0KNN accuracy 1.0Naive Bayes
Naïve Bayes is a probabilistic machine learning algorithm based on the Bayes Theorem, it is widely used in classification problems especially in natural language processing.
It assumes independence between predictors, in that the presence of a particular feature in a class is unrelated to the presence of any other feature.
Bayes Theorem
Assumption: All features are mutually independent
Probabilities: P(y|X)-posterior, P(x|y)-class conditional, P(y)- prior y, P(X) - prior x
Select the class with the highest Probability
Log prevents overflow problems from very small numbers after multiplications Prior Probability P(y): frequency
Class conditional probability P(x_i|y): create probability density functions using normal distributions of different means and variances
class NaiveBayes:
def fit(self, X, y):
# each class means and variances
n_samples, n_features = X.shape
self._classes = np.unique(y) #unique classes
n_classes = len(self._classes)
#init mean, var, priors
self._mean = np.zeros((n_classes, n_features), dtype=np.float64)
self._var = np.zeros((n_classes, n_features), dtype=np.float64)
self._priors = np.zeros(n_classes, dtype=np.float64)
for c in self._classes:
X_c = X[c==y] #filter rows with the class
self._mean[c, :] = X_c.mean(axis=0) #mean for rows
self._var[c, :] = X_c.var(axis=0)
self._priors[c] = X_c.shape[0] / float(n_samples) #class frequency
def predict(self, X):
y_pred = [self._predict(x) for x in X]
return y_pred
def _predict(self, x):
posteriors = []
for idx, c in enumerate(self._classes):
prior = np.log(self._priors[idx])
class_conditional = np.sum(np.log(self._pdf(idx, x)))
posterior = prior + class_conditional
posteriors.append(posterior)
return self._classes[np.argmax(posteriors)]
def _pdf(self, class_idx, x):
# probility density function
mean = self._mean[class_idx]
var = self._var[class_idx]
numerator = np.exp(- (x-mean)**2 / (2*var))
denominator = np.sqrt(2*np.pi*var)
return numerator/denominator
X, y = datasets.make_classification(n_samples=100, n_features=10, n_classes=2, random_state=5)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=5)
nb = NaiveBayes()
nb.fit(X_train, y_train)
predictions = nb.predict(X_test)
print("Naive Bayes Classifiction accuracy", accuracy(y_test, predictions))Naive Bayes Classifiction accuracy 0.9500000000000002Perceptron
A Perceptron is a linear machine learning algorithm for binary classification tasks. It is one of the simplest types of artificial neural networks, which is an important building block for deep learning.
A unit of a neural network, simulates the behavior of a cell. A cell receives an input signal and if it reaches a threshold it fires an output of 1 or 0.
Inputs > Weights > Net Input Function > Activation Function > Output
Linear Model
Activation Function
Unit step function
Approximation
First apply the linear model function, then the activation function
Perceptron Update Rule
For each training sample, update new weight as old weight plus the delta weight. The delta weight is given by (alpha * (y-predictedy) * samplex). Alpha is the learning rate between [0,1].
In calculating the delta weight, if there is no difference between the actual and the predicted there is no change. However if there is a difference (misclassification) the weight increases(1-0=1) or decrease (0-1=-1)
Perceptron works for linearly seperable classes, to improve try different activation functions such as sigmoid function and in updating weights replace the perceptron update rule with gradient descent
class Perceptron:
def __init__(self, lr=0.001, n_iters=1000):
self.lr = lr
self.n_iters = n_iters
self.activation_func = self._unit_step_func
self.weights = None
self.bias = None
def _unit_step_func(self, x):
return np.where (x >= 0, 1, 0)
def fit(self, X, y):
n_samples, n_features = X.shape
# init weights
self.weights = np.zeros(n_features)
self.bias = 0
y_ = [1 if i > 0 else 0 for i in y]
for _ in range(self.n_iters):
for idx, x_i in enumerate(X):
linear_output = np.dot(x_i, self.weights) + self.bias
y_predicted = self.activation_func(linear_output)
update = self.lr * (y_[idx] - y_predicted)
self.weights += update * x_i
self.bias += update
def predict(self, X):
linear_output = np.dot(X, self.weights) + self.bias
y_predicted = self.activation_func(linear_output)
return y_predicted
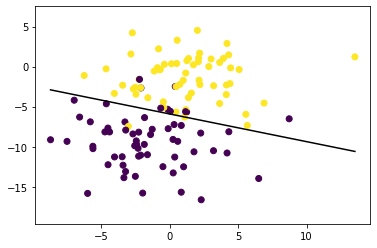
X, y = datasets.make_blobs(n_samples=150, n_features=2, centers=2, cluster_std=3.05, random_state=5)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=5)
p = Perceptron(lr=0.01, n_iters=1000)
p.fit(X_train, y_train)
predictions = p.predict(X_test)
print("Perceptron classification accuracy", accuracy(y_test, predictions))
fig = plt.figure()
ax = fig.add_subplot(1,1,1)
plt.scatter(X_train[:,0], X_train[:,1], marker='o', c=y_train)
x0_1 = np.amin(X_train[:, 0])
x0_2 = np.amax(X_train[:, 0])
x1_1 = (-p.weights[0] * x0_1 - p.bias) / p.weights[1]
x1_2 = (-p.weights[0] * x0_2 - p.bias) / p.weights[1]
ax.plot([x0_1, x0_2], [x1_1, x1_2], 'k') # decision boundary
ymin = np.amin(X_train[:,1])
ymax = np.amax(X_train[:,1])
ax.set_ylim([ymin-3, ymax+3])Perceptron classification accuracy 0.9666666666666667