Machine Learning From Scratch II
Decision Tree:
It has a tree-structure, where internal nodes represent the features of a dataset, branches represent the decision rules and each leaf node represents the outcome. It can be used for both regression and classification problems.
Entropy
Entropy is a measure of disorder or uncertainty and the goal of machine learning models is to reduce uncertainty. If the sample is completely homogeneous the entropy is zero and if the sample is an equally divided it has entropy of one
Given S=[0,0,0,0,0,1,1,1,1,1]
Information Gain
The information gain is the decrease in entropy after a dataset is split on an attribute. In the creation of a decision tree, we try to find an attribute that returns the highest information gain by having the most homogeneous branches.
Given S=[0,0,0,0,0,1,1,1,1,1], S1=[0,0,1,1,1,1,1], S2=[0,0,0]
Training process
- Starting at the top node, at each node select the best split based on the best information gain
- Loop over all features and over all thresholds (greedy search all feature values)
- Save the best split feature and split threshold at each node
- Build the tree recursively
- Apply some stopping criteria to stop growing e.g. maximum depth, minimum samples at node
- When we have a leaf node, store the most common class label of this node
Prediction Process
- Recursively traverse the tree
- At each node look at the best split feature of the test feature vector x and go on the left or right depending on x[feature_idx] <= threshold
- When we reach the leaf node we return the stored most common class label
from collections import Counter
def entropy(y):
counts = np.bincount(y) #count of occurences of class labels
prob_y = counts/len(y) # [3,6,9]/18 => 1/6, 1/3, 1/2
return -np.sum([p*np.log2(p) for p in prob_y if p> 0])
class Node:
def __init__(self, feature=None, threshold=None, left=None, right=None, *, value=None):
self.feature = feature
self.threshold = threshold
self.left = left
self.right = right
self.value = value
def is_leaf_node(self):
return self.value is not None
class DecisionTree:
def __init__(self, min_samples_split=2, max_depth=100, n_features=None):
self.min_samples_split = min_samples_split
self.max_depth = max_depth
self.n_features = n_features
self.root = None
def fit(self, X, y):
# grow the tree
self.n_features = X.shape[1] if not self.n_features else min(self.n_features, X.shape[1])
self.root = self._grow_tree(X, y)
def _grow_tree(self, X, y, depth=0):
n_samples, n_features = X.shape
n_labels = len(np.unique(y))
#stopping criteria
if (depth >= self.max_depth) \
or n_labels == 1 \
or (n_samples < self.min_samples_split):
leaf_value = self._most_common_label(y)
return Node(value=leaf_value)
#randomly select a few features
feat_idxs = np.random.choice(n_features, self.n_features, replace=False)
# greedy search
best_feat, best_thresh = self._best_criteria(X, y, feat_idxs)
left_idxs, right_idxs = self._split(X[:, best_feat], best_thresh)
# for each subset of rows find, best_criteria & split
left = self._grow_tree(X[left_idxs, :], y[left_idxs], depth+1)
right = self._grow_tree(X[right_idxs, :], y[right_idxs], depth+1)
return Node(best_feat, best_thresh, left, right)
def _best_criteria(self, X, y, feat_idxs):
# find the feature and threshold with the highest info gain
best_gain = -1
split_idx, split_thresh = None, None
for feat_idx in feat_idxs:
X_column = X[:, feat_idx]
thresholds = np.unique(X_column)
for threshold in thresholds:
gain = self._information_gain(y, X_column, threshold)
if gain > best_gain:
best_gain = gain
split_idx = feat_idx
split_thresh = threshold
return split_idx, split_thresh
def _information_gain(self, y, X_column, split_thresh):
# parent Entropy
parent_entropy = entropy(y)
# generate split
left_idxs, right_idxs = self._split(X_column, split_thresh)
if len(left_idxs) == 0 or len(right_idxs) == 0:
return 0
# weighted avearge child Entropy
n = len(y)
n_l, n_r = len(left_idxs), len(right_idxs)
e_l, e_r = entropy(y[left_idxs]), entropy(y[right_idxs])
child_entropy = (n_l/n) * e_l + (n_r/n) * e_r
ig = parent_entropy - child_entropy
return ig
def _split(self, X_column, split_thresh):
left_idxs = np.argwhere(X_column <= split_thresh).flatten()
right_idxs =np.argwhere(X_column > split_thresh).flatten()
return left_idxs, right_idxs
def _most_common_label(self, y):
counter = Counter(y)
most_common = counter.most_common(1)[0][0]
return most_common
def predict(self, X):
# traverse the tree
return np.array([self._traverse_tree(x, self.root) for x in X])
def _traverse_tree(self, x, node):
if node.is_leaf_node():
return node.value
if x[node.feature] <= node.threshold:
return self._traverse_tree(x, node.left)
return self._traverse_tree(x, node.right)
data = datasets.load_breast_cancer()
X, y = data.data, data.target
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=5)
clf = DecisionTree(max_depth=10)
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
acc = accuracy(y_test, y_pred)
print("Decision Tree accuracy:", acc
)Decision Tree accuracy: 0.9210526315789473Random Forest
Combines multiple trees each with a random subset of the data, predictions are made through majority voting by the trees. Multiple trees are better than a single tree in reducing overfitting and increasing accuracy
def bootstrap_sample(X, y):
n_samples = X.shape[0]
idxs =np.random.choice(n_samples, size=n_samples, replace=True)
return X[idxs], y[idxs]
def most_common_label(y):
counter = Counter(y)
most_common = counter.most_common(1)[0][0]
return most_common
class RandomForest:
def __init__(self, n_trees=100, min_samples_split=2, max_depth=100, n_features=None):
self.n_trees =n_trees
self.min_samples_split = min_samples_split
self.max_depth = max_depth
self.n_features = n_features
self.trees = []
def fit(self, X, y):
self.trees = []
for _ in range(self.n_trees):
tree = DecisionTree(min_samples_split=self.min_samples_split, max_depth=self.max_depth, n_features=self.n_features)
X_sample, y_sample = bootstrap_sample(X, y)
tree.fit(X_sample, y_sample)
self.trees.append(tree)
def predict(self, X):
tree_preds = np.array([tree.predict(X) for tree in self.trees])
tree_preds = np.swapaxes(tree_preds, 0, 1) #convert rows to cols
y_pred = [most_common_label(tree_pred) for tree_pred in tree_preds]
return np.array(y_pred)
data = datasets.load_breast_cancer()
X, y = data.data, data.target
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=5)
clf = RandomForest(n_trees=5) # try to optimize code for speed
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
acc = accuracy(y_test, y_pred)
print("Random Forest accuracy:", acc)Random Forest accuracy: 0.9298245614035088AdaBoost
Boosting is an ensemble modelling technique that improves the prediction power by converting a number of weak learners to strong learners.
The idea behind boosting algorithms is to build a model on the training dataset, then a second model is built to reduce the errors in the first model. This procedure is repeated until the errors are minimized, and the predictions are correct.
Error
Misclassified is given a higher weight in the next iteration
if error > 0.5, then error = 1- error
Weights
Update rule to make sure there is more weight for misclassified samples
Performance
Prediction
The sign of the sum of alpha times prediction. The better the learner the more it points to the positive or negative side.
Training Process
Initialize weights for each sample = .
for t in T:
- Train weak learner (find best feature and threshold)
-
Calculate the error for decision stump(1 feature tree split at a threshold)
- flip error and decision if error > 0.5
- Calculate
- Update weights:
class DecisionStump:
def __init__(self):
self.polarity = 1 # +/- 1
self.feature_idx = None
self.threshold = None
self.alpha = None
def predict(self, X):
n_samples = X.shape[0]
X_column = X[:, self.feature_idx]
predictions = np.ones(n_samples)
if self.polarity == 1:
predictions[X_column < self.threshold] =-1
else:
predictions[X_column > self.threshold] = -1
return predictions
class Adaboost:
def __init__(self, n_clf=5):
self.n_clf = n_clf
def fit(self, X, y):
n_samples, n_features = X.shape
# init weights
w = np.full(n_samples, (1/n_samples)) #fill array of n with x
self.clfs = []
# greedy search all features & threshold
for _ in range(self.n_clf):
clf = DecisionStump()
min_error = float('inf')
for feature_i in range(n_features):
X_column = X[:, feature_i]
thresholds = np.unique(X_column)
for threshold in thresholds:
pol = 1
predictions = np.ones(n_samples)
predictions[X_column < threshold] = -1
misclassified = w[y != predictions]
error = sum(misclassified)
if error > 0.5:
error = 1- error
pol = -1
if error < min_error:
min_error = error
clf.polarity = pol
clf.threshold = threshold
clf.feature_idx = feature_i
EPS = 1e-10 #prevent division by zero error
clf.alpha = 0.5 * np.log( (1-error) /(error+EPS))
predictions = clf.predict(X)
w *= np.exp(-clf.alpha * y * predictions) #exp(4) => 2.718**4
w /= np.sum(w)
self.clfs.append(clf)
def predict(self, X):
clf_preds = [clf.alpha * clf.predict(X) for clf in self.clfs]
y_pred = np.sum(clf_preds, axis=0)
y_pred = np.sign(y_pred) #negatives/positives are -1/1
return y_pred
data = datasets.load_breast_cancer()
X,y = data.data, data.target
y[y==0] = -1
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=5)
clf = Adaboost(n_clf=5)
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
acc = accuracy(y_test, y_pred)
print("Adaboost accuracy: ", acc)Adaboost accuracy: 0.9122807017543859Unsupervised Machine Learning
It is a type of machine learning in which models are trained using unlabeled dataset. The goal of unsupervised learning is to find the underlying structure of dataset, group that data according to similarities, and represent that dataset in a compressed format.
Principal Component Analysis (PCA)
PCA finds a new set of dimensions such that all the dimensions are orthogonal (linearly independent) and are ranked according to their variance. The transformed features should be linearly independent, with the their dimensionality reduced as only the dimensions with the highest importance are kept. The new dimensions should minimize the projection error (near projection line) and the projected points should have the maximum spread/variance.
Variance
The deviation of the data from the mean:
Covariance Matrix
The level to which two variables vary together
The autocovariance, is the covariation of a data point with another point of the same data.
Eigenvector & EigenValues
The eigenvector point in the direction of maximum variance, and the corresponding eigenvalues indicates the importance of its corresponding eigen vector.
When v is non-zero, we can solve the using the determinant.
A:matrix, v:eigenvector(direction that does not change) : eigenvalue(scale of the stretch), I: Identity matrix
Given a matrix
Calculating the determinant: (-6 - )(5-)- 3x4=0 => + - 42 = 0 => = -7 or 6
There are two possible eigenvalues =-7 and = 6. We can find their matching eigenvectors as follows.
After multiplying we get two equations:
Bring all to the left:
For eigenvalue =6, the eigenvector is any non-zero multiple of
PCA process
- Subtract the mean from X
- Calculate Cov(X, X)
- Calculate eigenvectors and eigenvalues of covarance matrix
- Sort the eigenvectors according to the eigenvalues in decreasing order
- Choose the first k eigenvectors and that will be the new k dimensions
- Transform the original n dimensional data points into k dimesions(Projection with dot product)
class PCA:
def __init__(self, n_components):
self.n_components = n_components
self.components = None
self.mean = None
def fit(self, X):
# mean
self.mean = np.mean(X, axis=0) #mean per row
X = X - self.mean
#covariance
cov = np.cov(X.T) #column covariance
#eigen vectors & eigenvalues
eigenvalues, eigenvectors = np.linalg.eig(cov)
#sort eigenvectors
eigenvectors = eigenvectors.T # returned as one column
idxs = np.argsort(eigenvalues)[::-1] #reverse a list
eigenvalues = eigenvalues[idxs]
eigenvectors = eigenvectors[idxs]
#store first n eigenvectors (n_components by X.shape[1])
self.components = eigenvectors[0:self.n_components]
def transform(self, X):
# project data
X = X - self.mean
return np.dot(X, self.components.T) #convert to columns
data = datasets.load_iris()
X = data.data
y = data.target
pca = PCA(2)
pca.fit(X)
X_projected = pca.transform(X)
print("Shape of X: ", X.shape)
print("Shape of transformed X:", X_projected.shape)
x1 = X_projected[:, 0]
x2 = X_projected[:, 1]
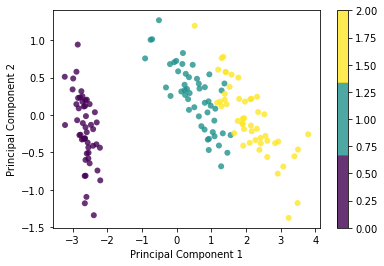
plt.scatter(x1, x2, c=y, edgecolor='none', alpha=0.8, cmap=plt.cm.get_cmap('viridis', 3))
plt.xlabel('Principal Component 1')
plt.ylabel('Principal Component 2')
plt.colorbar()
plt.show()Shape of X: (150, 4)
Shape of transformed X: (150, 2)Linear Discriminant Analysis
LDA is a feature reduction technique that projects a dataset onto a lower-dimensionality space with good class-separability by maximizing the component axes for class-separation.
PCA is unsupervised and finds the component axes that maximizes the data variance, LDA is a supervised method that finds axes that maximize the separation between classes.
Within class scatter matrix
Between class scatter matrix
Eigenvalue & Eigenvector
LDA Process
- Calculate
- Calculate eigenvalues of
- Sort the eigenvectors according to their eigenvalues in decreasing order
- Choose first k eigenvectors as the new k dimensions
- Transform the original n dimensional data points into k dimensional by projection with dot product
class LDA:
def __init__(self, n_components):
self.n_components = n_components
self.linear_discriminants = None #store eigenvectors
def fit(self, X, y):
n_features = X.shape[1]
class_labels = np.unique(y)
#S_W & S_B
mean_overall = np.mean(X, axis=0)
S_W = np.zeros((n_features, n_features))
S_B = np.zeros((n_features, n_features))
for c in class_labels:
X_c = X[y == c]
mean_c = np.mean(X_c, axis=0)
S_W += (X_c - mean_c).T.dot(X_c - mean_c)
# Between class
n_c = X_c.shape[0]
mean_diff = (mean_c - mean_overall).reshape(n_features, 1) #rows * col
S_B += n_c * (mean_diff).dot(mean_diff.T)
A = np.linalg.inv(S_W).dot(S_B) #matrix
eigenvalues, eigenvectors = np.linalg.eig(A)
eigenvectors = eigenvectors.T
idxs = np.argsort(abs(eigenvalues))[::-1]
eigenvalues = eigenvalues[idxs]
eigenvectors = eigenvectors[idxs]
self.linear_discriminants = eigenvectors[0:self.n_components]
def transform(self, X):
return np.dot(X, self.linear_discriminants.T)
data = datasets.load_iris()
X, y = data.data, data.target
# Project the data onto the 2 primary linear discriminants
lda = LDA(2)
lda.fit(X, y)
X_projected = lda.transform(X)
print('Shape of X:', X.shape)
print('Shape of transformed X:', X_projected.shape)
x1 = X_projected[:, 0]
x2 = X_projected[:, 1]
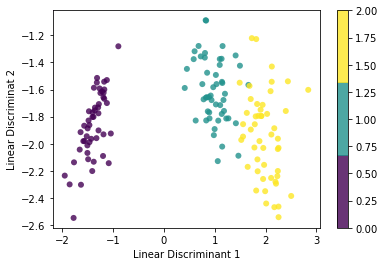
plt.scatter(x1, x2, c=y, edgecolor='none', alpha=0.8, cmap=plt.cm.get_cmap('viridis', 3))
plt.xlabel('Linear Discriminant 1')
plt.ylabel("Linear Discriminat 2")
plt.colorbar()
plt.show()KMEANS
An unsupervised machine learning method that clusters a dataset into k different clusters. Each sample is assigned to the cluster with the nearest mean.
Iterative optimization
- Initialize clusters centers randomly
- Assign points to the nearest cluster center (centroid)
- Repeat until convergence to update cluster labels
- Set the center to the mean of each cluster
- Repeat until convergence to update cluster centroids
Euclidean distance
Get the distance between two feature vectors
from sklearn.datasets import make_blobs
np.random.seed(42)
import matplotlib.pyplot as plt
def ecudlidean_distance(x1, x2):
return np.sqrt(np.sum((x1-x2)**2))
class KMeans:
def __init__(self, K=5, max_iters=100, plot_steps=False):
self.K = K
self.max_iters = max_iters
self.plot_steps = plot_steps
# list of sample indices for each cluster
self.clusters = [[] for _ in range(self.K)]
# mean feature vector for each cluster
self.centroids = []
def predict(self, X):
self.X = X
self.n_samples, self.n_features = X.shape
# init centroids
random_sample_idxs = np.random.choice(self.n_samples, self.K, replace=False)
self.centroids = [self.X[idx] for idx in random_sample_idxs]
# optimization
for _ in range(self.max_iters):
# update clusters
self.clusters = self._create_clusters(self.centroids)
if self.plot_steps:
self.plot()
# update centroids
centroids_old = self.centroids
self.centroids = self._get_centroids(self.clusters)
if self.plot_steps:
self.plot()
# check if converged
if self._is_converged(centroids_old, self.centroids):
break
# return cluster labels
return self._get_cluster_labels(self.clusters)
def _get_cluster_labels(self, clusters):
labels = np.empty(self.n_samples) #empty creates array of set shape
for cluster_idx, cluster in enumerate(clusters):
for sample_idx in cluster:
labels[sample_idx] = cluster_idx
return labels
def _create_clusters(self, centroids):
clusters = [[] for _ in range(self.K)]
for idx, sample in enumerate(self.X):
centroid_idx = self._closest_centroid(sample, centroids)
clusters[centroid_idx].append(idx)
return clusters
def _closest_centroid(self, sample, centroids):
distances = [ euclidean_distance(sample, point) for point in centroids]
closest_idx = np.argmin(distances)
return closest_idx
def _get_centroids(self, clusters):
centroids = np.zeros((self.K, self.n_features))
for cluster_idx, cluster in enumerate(clusters):
cluster_mean = np.mean(self.X[cluster], axis=0)
centroids[cluster_idx] = cluster_mean
return centroids
def _is_converged(self, centroids_old, centroids):
distances = [euclidean_distance(centroids_old[i], centroids[i]) for i in range(self.K)]
return sum(distances) == 0
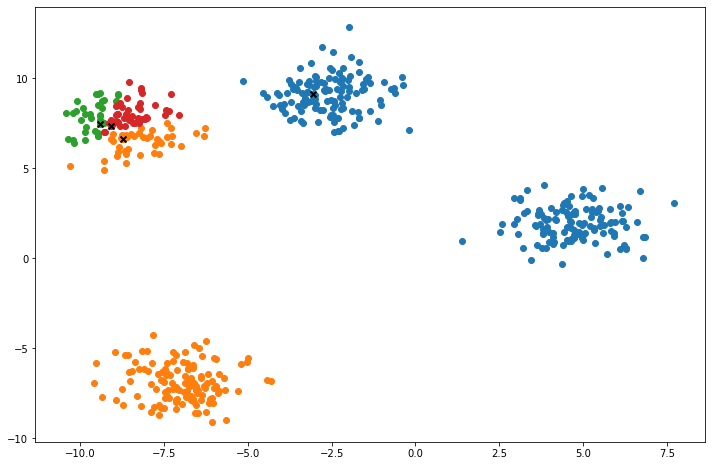
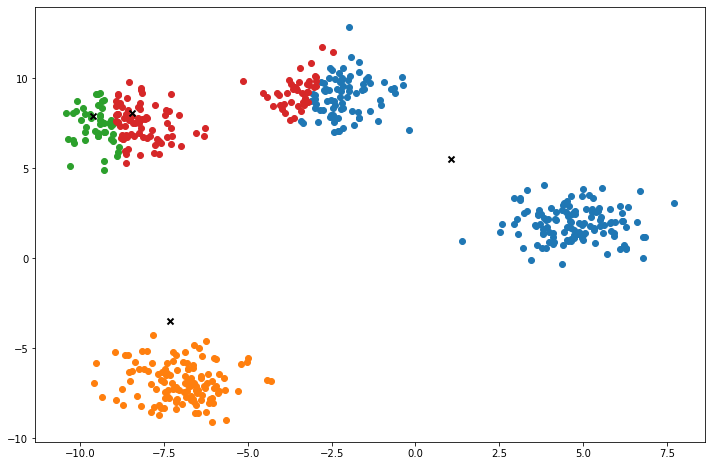
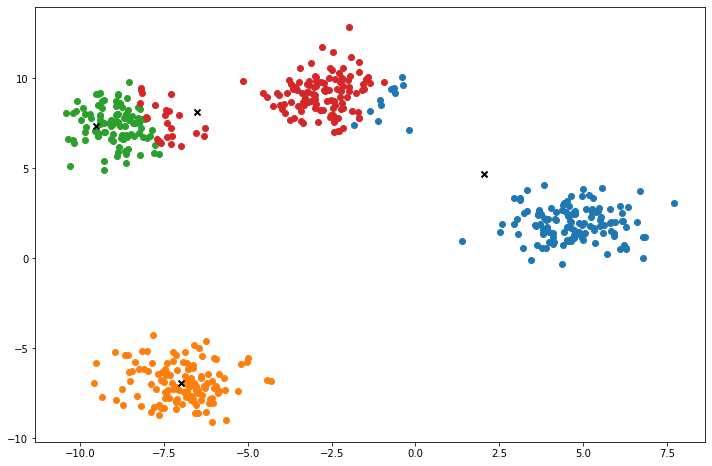
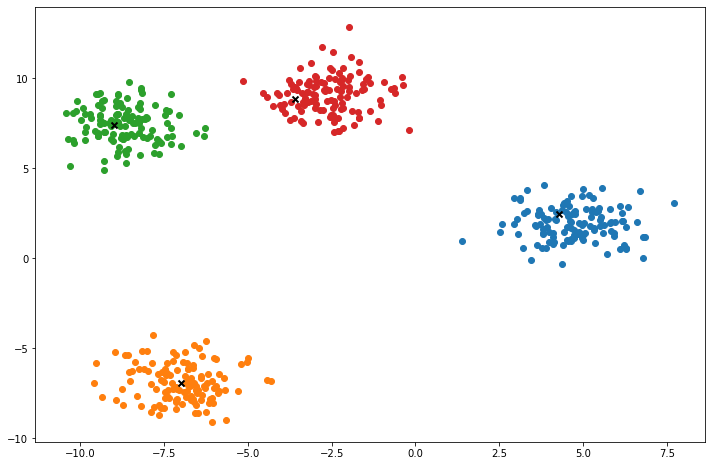
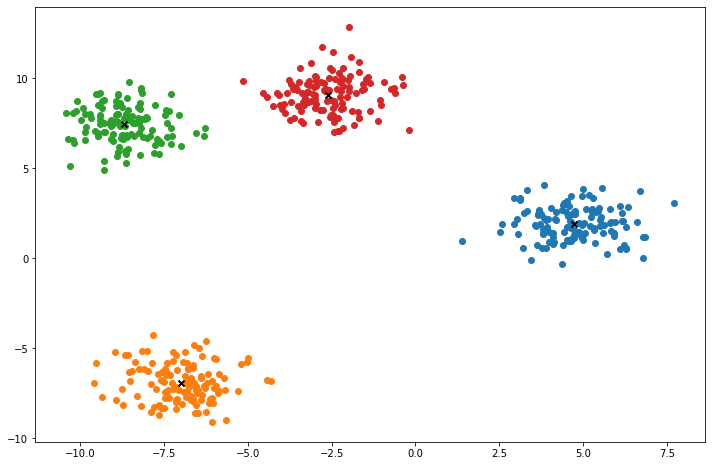
def plot(self):
fig, ax = plt.subplots(figsize=(12, 8))
for i, index in enumerate(self.clusters):
point = self.X[index].T
ax.scatter(*point)
for point in self.centroids:
ax.scatter(*point, marker='x', color='black', linewidth=2)
plt.show()
X, y = make_blobs(centers=4, n_samples=500, n_features=2, shuffle=True, random_state=5)
clusters = len(np.unique(y))
print("clusters:", clusters)
k = KMeans(K=clusters, max_iters=150, plot_steps=True)
y_pred = k.predict(X)Cluster 4Machine Learning in practice
Machine learning is applied in pattern and image recognition, web search results, fraud detection, credit scoring, and in serving adverts on webpages. In conclusion, machine learning can produce accurate results and analysis from big data by developing fast and efficient algorithms for real-time data processing.